Submission requirements
- A single chain protein structure in PDB format
- Contains metal ion(s) one of the following residue codes: MO, MOO, 4MO, 6MO, MOS, MG, 3NI, NI, ZN, MGF, MN3, MN, CO, OFE, FE2, FEO, FE, FES, CU, C2O, CUA, CU1, 3CO
- Metal sites with more than 5 metal ions will not receive predictions
- Protein structures with more than 1000 residues will not receive predictions from the webserver, but these structures can be used with the downloadable MAHOMES II
Detailed overview
MAHOMES II (Metal Activity Heuristic Of Metal and Enzyme Sites 2) is a structure-based machine learning tool for predicting protein bound metal ions to be enzymatic or non-enzymatic. The MAHOMES II website performs two tasks 1) use an automated feature pipeline to calculate the necessary structure-based features and 2) make enzyme or non-enzyme predictions using MAHOMES II.
This page covers relevant details for usage of the MAHOMES II website. Details about MAHOMES II’s methods can be found in publications (Feehan, Franklin, & Slusky, 2021). MAHOMES II uses a gradient boosting classifier from scikit-learn(Pedregosa et al., 2011), which can be read about at https://scikit-learn.org/stable/modules/ensemble.html#gradient-boosting.
MAHOMES II output
Users will receive an email, when a submitted job has finished running, which will include a link to that job's results. The outputs page will contain a row for each metal site, defined as four or fewer metal ion(s) within 5 Å of each other, or for an input structure that failed the automated feature calculation process.
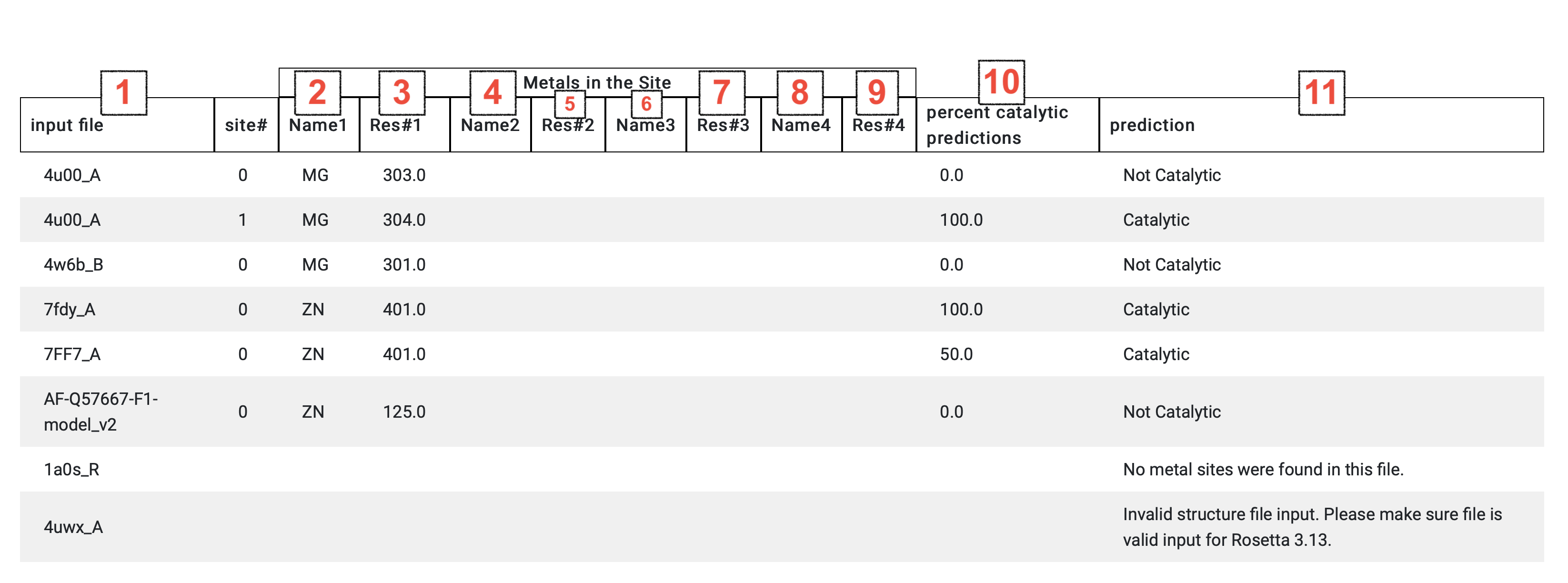
Site results include (1) the user’s file name for the site’s structure, the residue code and the residue number for the site’s (2,3) first metal ion, (4,5) second metal ion, (6,7) third metal ion, and (8,9) fourth metal ion when relevant.
MAHOMES II used ten different gradient boosting ML models, which use different random seeds for training. Results for each site display (10) the percent of these models that predicted the site to be enzymatic and (11) the final enzyme or non-enzyme prediction. For structures that failed the feature calculation process, an explanation will be given in place of a prediction.
Automated feature calculation pipeline
Physicochemical features are calculated covering five categories – Rosetta energy terms, pocket void, pocket lining, electrostatics, and coordination geometry. These calculations use Rosetta (Alford et al., 2017), BLUUES (Fogolari et al., 2012), pdb2pqr (Jurrus et al., 2018), FindGeo (Andreini, Cavallaro, & Lorenzini, 2012), and GHECOM(Kawabata, 2019). Due to the use of these third-party tools, only the final prediction made by MAHOMES II is made accessible to users. However, in the event that the automated feature process fails, feedback is given in the results to tell users what went wrong so that they may attempt to make necessary adjustments. Feedback messages are detailed below.
Invalid input file: Metalloprotein structure files are first input into Rosetta to create a uniform .pdb output for further processing. “Invalid input file” means that Rosetta was unable to use this file. The main reason for this issue is improper PDB format. For other potential causes, scoring the file with Rosetta 3.13 should create a Rosetta crash report that details the issue.
No metal ions in structure: None of the relevant metal ion codes were found in the structure file. Note that MAHOMES II only works for metal ions listed in the submission requirements.
To many metals in the site: Feature calculations did not proceed because this site had more than four metals. Sites include any metal ion within 5 Å of at least one metal ion in the site. We use this limit because of issues with feature calculations like those that calculate distances from the sites geometric center.
Unknown feature calculation failure: Congratulations on finding a new bug. We can be contacted by emailing mahomes@ku.edu to see if this is an issue that we can resolve. Alternatively, the code for the automated feature pipeline and MAHOMES II can be downloaded.
Placing a metal site with known coordination residues
Our performance evaluation of MAHOMES II included a test of metal sites on computationally generated structures. The original computationally generated structures included only the twenty canonical residues. Using coordinating residues identified on UniProt, we placed metals relevant metal binding sidechain atoms. The script we used for metal binding site placement, addMetalIon.py, is available for download with the rest of our MAHOMES II on GitHub. Additionally, the metalloprotein structures and enzyme/non-enzyme labels are available on Zenodo.
References
-
Feehan, R, Copeland, M, Franklin, MW, Slusky, JSG. (2023).
MAHOMES II: A webserver for predicting if a metal binding site is enzymatic.
Protein Science. 32(4): e4626.doi: 10.1002/pro.4626 - Alford, R. F., Leaver-Fay, A., Jeliazkov, J. R., O’Meara, M. J., DiMaio, F. P., Park,
H., . . . Gray, J. J. (2017). The Rosetta All-Atom Energy Function for
Macromolecular Modeling and Design. Journal of Chemical Theory and
Computation, 13(6), 3031-3048. doi:10.1021/acs.jctc.7b00125 - Andreini, C., Cavallaro, G., & Lorenzini, S. (2012). FindGeo: a tool for
determining metal coordination geometry. Bioinformatics, 28(12),
1658-1660. doi:10.1093/bioinformatics/bts246 - Feehan, R., Franklin, M. W., & Slusky, J. S. G. (2021). Machine learning
differentiates enzymatic and non-enzymatic metals in proteins. Nature
Communications, 12(1), 3712. doi:10.1038/s41467-021-24070-3 - Fogolari, F., Corazza, A., Yarra, V., Jalaru, A., Viglino, P., & Esposito, G.
(2012). Bluues: a program for the analysis of the electrostatic
properties of proteins based on generalized Born radii. BMC
Bioinformatics, 13(4), S18. doi:10.1186/1471-2105-13-S4-S18 - Jurrus, E., Engel, D., Star, K., Monson, K., Brandi, J., Felberg, L. E., . . . Baker,
N. A. (2018). Improvements to the APBS biomolecular solvation
software suite. Protein Science, 27(1), 112-128.
doi:10.1002/pro.3280 - Kawabata, T. (2019). Detection of cave pockets in large molecules: Spaces
into which internal probes can enter, but external probes from outside
cannot. Biophysics and physicobiology, 16, 391-406.
doi:10.2142/biophysico.16.0_391 - Pedregosa, F., Varoquaux, G., Gramfort, A., Michel, V., Thirion, B., Grisel, O., .
. . Dubourg, V. (2011). Scikit-learn: Machine learning in Python. The
Journal of Machine Learning Research, 12, 2825-2830.